Potential applications of IHC to aid pathological assessment of SCLC
Authors:
Dr. Anna Keogh1
Dr. Teodora Radonic2
Prof. Stephen P. Finn1
1 Department of Pathology, St. James Hospital, Saint James’ (part of Phoenix Park), Dublin
E-Mail: akeogh5@tcd.ie
2 Department of Pathology,
Amsterdam University Medical Centre
E-Mail: t.radonic@amsterdamumc.nl
Small-cell lung cancer (SCLC) is an aggressive tumor type with dismal prognosis. In this article, we will discuss the recent progress in understanding the biology of SCLC and how application of immunhistochemistry (IHC) may enable the development of novel diagnostic, prognostic, and predictive biomarkers to drive clinical decisions for these aggressive tumours in the future.
Keypoints
-
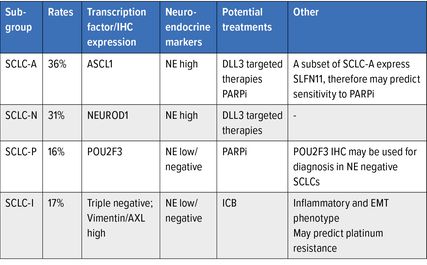
SCLC has been divided into four subtypes as defined by the expression of distinct transcriptional regulators which are detectable by IHC.
-
The four subtypes, SCLC-A, SCLC-N, SCLC-P and SCLC-I represent 36%, 31%, 16% and 17% respectively. They have distinct biological attributes and in preclinical studies have been shown to have different therapeutic vulnerabilities.
-
SCLC-I demonstrates a mesenchymal phenotype and may exhibit chemoresistance. This subtype also exhibits infiltration of immune cells and checkpoints and may predict ICB benefit.
-
SLFN11 (expressed in a subset of SCLC-A) as well as SCLC-P have been shown to predict response to PARPi in patients and cell line models of SCLC.
-
Identifying these subgroups, which can easily be done using IHC, will hopefully enable a more personalized approach to treatment of patients with SCLC.
For almost 80 years immunohistochemistry (IHC) has been a valuable tool for pathologists. IHC has a vast number of applications including its use as a prognostic marker in cancer, for diagnosis of tumours of uncertain histogenesis, and prediction of response to therapy. Moreover, IHC, when properly validated with appropriate controls, is a relatively simple technique to perform that is highly reproducible and reliable, as well as being low cost.
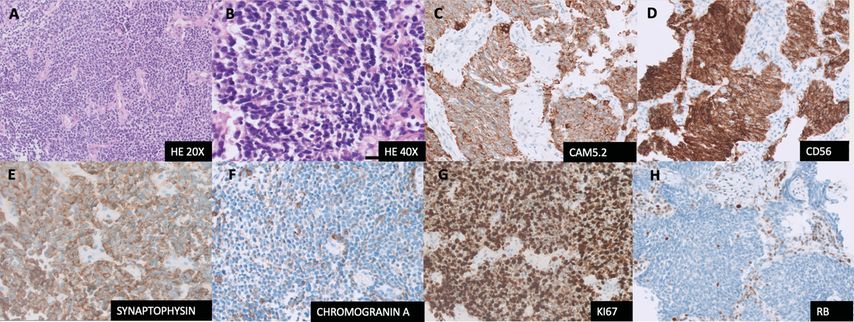
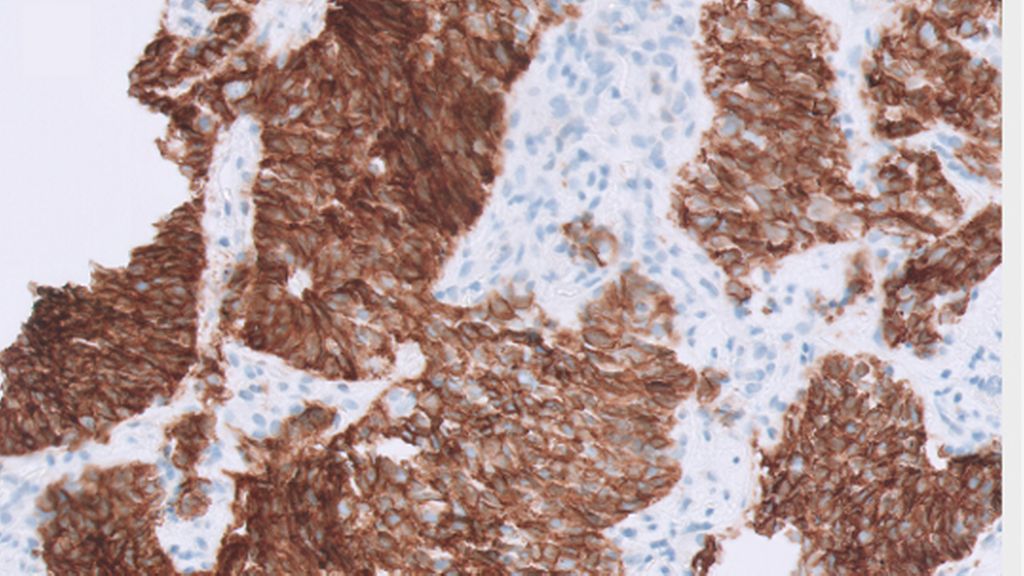
Small-cell lung cancer (SCLC) is an aggressive neuroendocrine malignancy with a dismal prognosis accounting for 15% of all lung cancers.1 Histology of SCLC is characterized by relatively small monomorphic nuclei with uniform dark chromatin pattern, inconspicuous nucleoli and scant cytoplasm (Fig. 1 A, B). IHC used for SCLC is for diagnostic purposes only, being positive for keratin (characteristic dot-like positivity) and at least one of the neuroendocrine (NE) markers CD56, synaptophysin and chromogranin A (Fig. 1D-F). In addition, Ki-67 proliferative index is high (>50%; Fig 1G).2 Currently, there are no biomarkers available to guide clinical decisions in SCLC. Multiple recent studies have identified molecular subsets within SCLC based on differential gene expression and methylation profiles.
Figure 1 (A-H): Small cell lung cancer. A, B: HE image of SCLC at 20X and 40X respectively. C: CAM 5.2 with dot-like pattern of cytoplasmic positivity. D; CD56, strong positivity. E: Synaptophysin, diffusely positive. F: Chromogranin A, dot like positivity in cytoplasm, pathognomonic of SCLC. G: Ki-67, demonstrates high proliferation index >80%. H: RB, loss of nuclear staining in the tumour nuclei, note the wild type pattern of staining in the background stromal cells
Emerging subgroups of SCLC
SCLCs are genomically homogenous compared to NSCLCs, characterised almost always by homozygous functional inactivation of RB and TP53.1 Complete loss of RB in IHC (Fig. 1H) or a mutant expression of p53 IHC can be helpful in confirming the diagnosis of SCLC in difficult cases, although validation studies for daily clinical utility are scarce.
Genomic subtyping studies looking at the tumour mutational landscape have failed to identify recurrent, actionable oncogenic mutations. High tumour mutational burden, characteristic of SCLC, did not result in meaningful treatment options for SCLC patients.3 However, recent studies have identified the expression of relatively distinct transcription signatures that permit classification of SCLCs into four biologically distinct subtypes.4,5
In one of the most recent studies Gay et al. applied non-negative matrix factorization (NMF) to previously published RNAseq data from 81 surgically resected, mostly limited stage SCLC and identified four groups that were defined primarily by master transcriptional regulators:
-
those with high expression of «Achaete-scute homolog 1» (ASCL1; SCLC-A),
-
those with high «Neurogenic differentiation 1» (NEUROD1; SCLC-N),
-
those with high «POU class 2 homeobox 3» (POU2F3; SCLC-P), and
-
a fourth group with low expression of all three transcription factor signatures but with high infiltration of immune cells and checkpoints, termed inflamed SCLC (SCLC-I).5
The former three had been previously described4 and the latter being a newly defined group. SCLC-A, SCLC-N, SCLC-P and SCLC-I were identified in 36%, 31%, 16% and 17% of SCLCs, respectively.5
These four groups have distinct NE marker expression patterns with SCLC-A and SCLC-N having high expression of NE markers and SCLC-I and SCLC-P having low/no expression (Tab. 1).
SCLC-I group was characterised as having the most mesenchymal phenotype, expressing low levels of e-cadherin (epithelial marker) and higher levels of vimentin (mesenchymal marker) indicative of epithelial mesenchymal transition (EMT). Interestingly, this study also described the plasticity of SCLC subtypes and ability to subclass switch from other SCLC subtypes to SCLC-I after first line chemotherapy, most probably the mechanism of chemoresistance in SCLC (not discussed further in this article).6
Subsequently, these gene expression signatures were validated using IHC for ASCL1 (SCLC-A), NeuroD1 (SCLC-N) and POU2F3 (SCLC-P).5,7 The SCLC-I group stained negative for all three IHC (triple negative) but was high in vimentin IHC.
In another study, these four subtypes were validated on a single cell level and a novel potential subtype emerged with high phospholipase C gamma 2 (PLCG2). Clinically, this clone was characterized by a high metastatic potential and could show overlap with the four subtypes. Moreover, a prominent immunosuppressive environment and T-cell exhaustion were associated with PLCG2 clone.8
What is the significance of detection of these groups? Each of these groups have been shown to have distinct biological attributes and, in preclinical studies these subtypes have been shown to have different therapeutic vulnerabilities (Tab. 1) and therefore may be used to determine clinical decisions for these patients in the future.
IHC as a diagnostic biomarker
Traditionally, diagnosis of SCLC has been made on morphology alone. However, IHC is widely used in clinical practice, increasing the accuracy of diagnosis.9 Histologically, the differential diagnosis of SCLC includes other neuroendocrine carcinomas, basaloid squamous cell carcinoma, small round cell sarcomas, and lymphoma. The clinical course and treatments vary widely across these malignancies and therefore accurate diagnosis is essential.
Positive staining for at least one of the three NE markers (synaptophysin, chromogranin and CD56) occurs in 90–95% of cases, however 5–10% are negative for all three.10 SCLC-P with POU2F3 IHC has been shown to be expressed in SCLC that are NE low/-.10 Although the occurrence of tumours with SCLC morphology and negativity for all NE markers is rare, it creates a diagnostic dilemma usually with large IHC panels needed to exclude other differentials, often delaying diagnosis. The addition of POU2F3 IHC therefore would aid in a more efficient diagnostic pathway for these NE low/- SCLCs. However, specificity of POU2F3 for SCLC is yet to be explored in future studies.
IHC as a prognostic biomarker
Classification of SCLC into these four biological subtypes has not been translated to large cohorts of SCLC patients in order to investigate their clinical significance. The four subgroups might prove to have prognostic value for these patients. Most SCLC patients present with metastatic disease1 and the less prevalent limited stage SCLC might therefore be a separate biological entity, clinically and genetically. Future investigation of the described SCLC subgroups might reveal prognostically relevant subtypes, however, for now, SCLC subtype behaviour in a clinical context is still not clarified.
Furthermore, IHC for PLCG2, which has been shown to be differentially expressed in sub-populations within SCLC tumours, when high is linked to metastatic ability,8 and may be useful in the future as a prognostic biomarker.
IHC as a predictive biomarker
Currently, there are no biomarkers by IHC or otherwise to guide therapeutic clinical decision making in SCLC.
Addition of atezolizumab to standard first line SCLC treatment improved the progression-free and overall survival in the IMpower133 trial, although with modest hazard ratios and relatively small group of patients showing benefit of immune checkpoint blockade (ICB) addition.11 Programmed cell death 1 ligand 1 (PD-L1) is rarely expressed in SCLC12 and was not helpful in predicting response to immunotherapy and therefore, the recent approval for first-line immunotherapy did not require PD-L1 IHC.11 However, when the gene expression data from the IMpower133 trial patients were analysed for the described four transcriptional subsets, SCLC-I subtype was enriched in patients with a significant OS benefit, suggesting that SCLC-I subtype may predict ICB benefit. However, there is no single IHC specific for the SCLC-I subtype which is rather characterized by a diffuse infiltration of immune cells and EMT; thus strong vimentin positivity might be indicative of this subtype. However, additional studies might identify the more precise IHC approach to identify the SCLC-I subtype. Other potential emerging predictive biomarkers include «Delta-like canonical notch ligand 3» (DLL3) and Schlafen-11 (SLFN11).
DLL3 has been shown to be highly and specifically expressed in SCLC and negative in most normal tissues making it an attractive potential therapeutic target. DLL3 plays a crucial role in Notch signalling pathway as a downstream target of ASCL1 and is therefore enriched in the SCLC-A ad SCLC-N subtypes.7 Despite the failure of DLL3 specific antibody-drug conjugate rovalpituzumab tesirine (ROVA-T) in phase II and III studies to demonstrate improvement of OS,13 DLL3 remains an attractive target in SCLC due to its high expression levels and specificity. Novel therapeutic strategies, namely anti-DLL3 bispecific T-cell engager are in development.14 DLL3 can be readily detected by IHC in SCLC and might emerge as a predictive biomarker for selecting patients for the treatment with DLL3 targeted therapies in the future.
Another potential therapeutic strategy in SCLC are DNA damage-repair proteins, such as poly [ADP-ribose] polymerase (PARP) inhibitors (PARPi). PARPi have shown promising results in terms of objective response rates of approximately 40% when used in combination with temozolomide for patients with relapsed SCLC,15 although progression free survival and OS did not show significant differences in this randomised phase II trial.15 This has led to investigations into rational patient selection for tumours with PARPi sensitivity who may have survival benefit. Schlafen-11 (SLFN11) has been identified as an emerging predictive biomarker of PARPi in SCLC pre-clinical models.6 Patients with SLFN11 positive tumours in IHC were found to have significantly improved PFS and OS compared with the those who had SLFN11 negative SCLCs when treated with PARPi.16 In the preclinical models SCLC-P subtype has also been shown to be sensitive to PARPi, independent of SLFN11 expression.17
Together these data indicate a potential role of POU2F3 and SLFN11 IHC in patient selection for PARPi. SLFN11 IHC has consistently been shown to be a reliable method for detection of expression in SCLC as well as multiple other cancers.16,18,19 IHC shows prominent nuclear staining20 with any nuclear staining regarded as having positive expression and complete loss of staining being negative21 making it an easily interpretable marker.
Taken together, despite the morphological homogeneity of SCLC under the microscope, novel developments indicate biological heterogeneity of SCLC. SCLC has traditionally been a graveyard of clinical trials without novel treatment options beyond the platinum based chemotherapy. Perhaps, a more personalised approach is warranted for development of novel treatment options. Future prospective trials are needed to elucidate the clinical relevance of the novel transcriptional subsets in SCLC.
To put it in a nutshell
Recent studies have led to a deeper understanding of the biology of SCLC. Currently there are four subtypes of SCLC identified based expression of different transcription factors. Identifying these subgroups will hopefully allow for a more personalized approach to the treatment of patients with SCLC. IHC is a reliable and cost-effective tool that can be easily implemented and may be used to develop novel diagnostic, prognostic, and predictive biomarkers to aid in the pathologic assessment and guide clinical decisions for these aggressive tumours in the future.
Literature
1 Rudin CM et al.: Nat Rev Dis Primers 2021; 7(1): 3 2 Pelosi G et al.: J Thorac Oncol 2014; 9(3): 273-84 3 Paz-Ares L et al.: Ann Oncol 2019; 30: 917-8 4 Rudin CM et al.: Nat Rev Cancer 2019; 19(5): 289-97 5 Gay CM et al.: Cancer Cell 2021; 39(3): 346-60 e7 6 Allison Stewart C et al.: Oncotarget 2017; 8(17): 28575-87 7 Baine MK et al.: J Thorac Oncol 2020; 15(12): 1823-35 8 Chan JM et al.: Cancer Cell 2021; 39(11): 1479-96.e18 9 Organisation WH: 2021. 5th ed. Lyon (France): WHO Classification of Tumours Editorial Board 2021 10 Rekhtman N: Mod Pathol 2022; 35(Suppl 1): 36-50 11 Horn L et al.: N Engl J Med 2018; 379(23): 2220-9 12 Schultheis AM et al.: Eur J Cancer 2015; 51(3): 421-6 13 Matsuo K et al.: Cancer Sci 2021; 112(8): 2984-92 14 Hipp S et al.: Clin Cancer Res 2020; 26(19): 5258-68 15 Farago AF et al.: Cancer Discov 2019; 9(10): 1372-87 16 Pietanza MC et al.: J Clin Oncol 2018; 36(23): 2386-94 17 Knelson EH et al.: Cancers (Basel) 2021; 13(4) 18 Lok BH et al.: Clin Cancer Res 2017; 23(2): 523-35 19 Takashima T et al.: Virchows Arch 2021; 478(3): 569-79 20 Gardner EE et al.: Cancer Cell 2017; 31(2): 286-99 21 Zhang B et al.: Br J Cancer 2021; 125(10): 1333-40
Das könnte Sie auch interessieren:
Erhaltungstherapie mit Atezolizumab nach adjuvanter Chemotherapie
Die zusätzliche adjuvante Gabe von Atezolizumab nach kompletter Resektion und adjuvanter Chemotherapie führte in der IMpower010-Studie zu einem signifikant verlängerten krankheitsfreien ...
Highlights zu Lymphomen
Assoc.Prof. Dr. Thomas Melchardt, PhD zu diesjährigen Highlights des ASCO und EHA im Bereich der Lymphome, darunter die Ergebnisse der Studien SHINE und ECHELON-1
Aktualisierte Ergebnisse für Blinatumomab bei neu diagnostizierten Patienten
Die Ergebnisse der D-ALBA-Studie bestätigen die Chemotherapie-freie Induktions- und Konsolidierungsstrategie bei erwachsenen Patienten mit Ph+ ALL. Mit einer 3-jährigen ...